Welcome to the PeakForest collaborative portal and spectral databases Hub for Metabolomics community.

PeakForest is a project funded by MetaboHUB and INRAE.
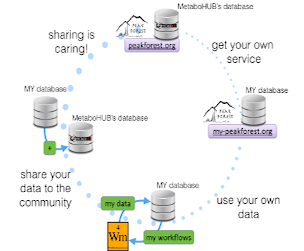
PeakForest framework
- Public repository of PeakForest is hosted on gitHub at https://github.com/peakforest with last APIs releases.
- Public repository on DockerHub
- General WebServices documentation (based on Alpha instance)
- Install documentation (from scratch or docker mode)
- Standard operating procedures
- And short HOWTO tutorials
PeakForest Demo

Interested by the PeakForest project? Please visit our demo instance and don't hesitate to ask a account to test more advanced functionalities (user or curator).
Citing the PeakForest project
1. Paulhe, N., Canlet, C., Damont, A. et al. PeakForest: a multi-platform digital infrastructure for interoperable metabolite spectral data and metadata management. Metabolomics 18, 40 (2022). https://doi.org/10.1007/s11306-022-01899-3
Public PeakForest Servers
Public servers are opened to the metabolomics academic community.
Ask your own PeakForest instance
PeakForest is a project hosting several instances.
Private PeakForest Servers
Private servers are PeakForest instances for closed consortium.